Introduction to the genetic aspect of periodontal diseases
Periodontitis may have a slow (Grade A), moderate (Grade B) or rapid (Grade C) rate of progression (see “Classification of periodontal and peri-implant diseases and conditions”). Grade A/B periodontitis has a slow rate of disease progression which is consistent with the presence of local factors; whereas, Grade C periodontitis is characterized by the rapid periodontal breakdown which is inconsistent with the presence of local factors. So, why a patient who is maintaining an adequate oral hygiene has periodontal breakdown? This question can be answered on the basis of multifactorial etiology of periodontal diseases of which genetics is an important component.
To understand the genetic connection of periodontal diseases, first, we need to understand that host response against plaque bacterial challenge leads to the activation of inflammatory and immunological cascades, which result in the destruction of connective tissue and bone. Host response has been divided into two types, namely innate and adaptive immune response (“Basic concepts in immunity and inflammation”). The adaptive immune response is specifically “tailored” against the microbial challenge. It involves the production of specific antibodies by B-lymphocytes directed against particular oral microorganisms and the cell-mediated immune response that involves cytotoxic T-lymphocytes directed against the virus or bacteria-infected cells. This inflammatory and immunological response have been shown to be under genetic control 1. Any specific variation in this immune response due to genetic recombination may affect the normal immune response, making certain individuals respond to the microbial challenge in a different way.
Historical background
Research to find out the genetic factors involved in the progression of periodontal diseases started with the classical study by Löe et al. (1986) 2 on Sri Lankan tea workers. In their classic study on the natural history of periodontitis, they found that among individuals with poor oral hygiene and no access to dental care, some developed the disease at a rapid rate, whereas others experienced little or no disease. In this study, it was appreciated that some unrecognized environmental factors or some individual differences in the susceptibility to disease are present in the population under study. During the same period, the importance of genetic variations in determining the development and severity of periodontal disease, with genetic influences accounting for as much as ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Chromosome nomenclature
Cytogenetics is the study of chromosome number, structure, function, and behavior in relation to gene inheritance, organization and expression. The centromere divides chromosomes into a short arm (p; petit) and a long arm (q). There are four types of chromosomes: telocentric, acrocentric, submeta-centric and metacentric. This classification is based on the position of the centromere.
- Telocentric: Centromere is on one end, so no p arm is present
- Acrocentric: Centromere is very near to the end, so chromosome has a very small p arm
- Submetacentric: Centromere is close to the middle of the chromosome, so p arm is just a little smaller than q arm
- Metacentric: Centromere in exactly at the middle of the chromosome, so the p and the q arms are exactly of the same length.
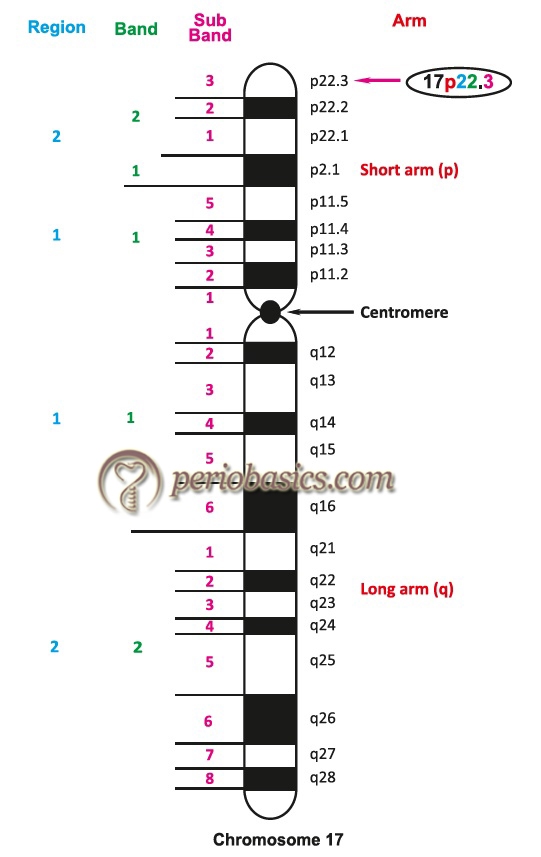
Each chromosome arm is divided into regions based on landmarks (consistent and distinct morphological area on a chromosome). The regions immediately adjacent to the centromere are designated as “1” (p1 and q1). Region numbers increase distally as we move across the arm from the centromere. Regions are divided into bands and bands into sub-bands, for example, 17p22.3 indicates chromosome 17, short arm region 2, band 2, sub-band 3. It is read as 17 p two-two point three, and not as 17 p twenty-two point three.
Terminologies
Before going into details of the topic, let’s first go through the basic terminologies used in the chapter,
Chromosome:
One of the threadlike “packages” of genes and other DNA in the nucleus of a cell. Different kinds of organisms have different numbers of chromosomes. Humans have 23 pairs of chromosomes, 46 in all: 44 autosomes and two sex chromosomes.
Autosomes:
Any chromosome other than a sex chromosome is called as autosome. Humans have 22 pairs of autosomes.
Sex chromosomes:
Humans have a total of 23 pairs of chromosomes, out of which one pair which determines the sex of the individual is called, the sex chromosome pair.
Gene:
It is the functional and physical unit of heredity passed from parents to offspring. Genes are pieces of DNA, and most genes contain the information for making a specific protein.
Exons and Introns:
Exon is a segment of a gene that contains information used in coding for protein synthesis. Genetic information within genes is discontinuous, split among the exons that encode for messenger RNA and absent from the DNA sequences in between, which are called introns. Genetic splicing, catalyzed by enzymes, results in the final version of messenger RNA, which contains only genetic information from the exons.
Recessive:
A genetic disorder that appears only in patients who have received two copies of a mutant gene, one from each parent.
Dominant:
A gene that almost always results in a specific physical characteristic, for example, a disease, even though the patient’s genome possesses only one copy. With a dominant gene, the chance of passing on the gene (and therefore the disease) to children is 50-50 in each pregnancy.
Penetrance:
The regularity with which an allele is expressed in a person who carries it. If an allele always produces its effect on the phenotype, it is fully penetrant. If an allele produces its effect less frequently than 100% of the time, it is not fully penetrant.
Genotype:
The genetic characteristics that determine the structure and function of an organism, i.e. the genetic constitution of an organism. When applied to a single gene locus, the term genotype is used to distinguish one allele, or a combination of alleles, from the others. The genotype of an organism determines its phenotype.
Phenotype:
The observable traits or characteristics of an organism, for example, hair color, weight, or the presence or absence of a disease. Phenotypic traits are not necessarily genetic.
Haplotype (haploid genotype):
A haplotype is a group of genes within an organism that was inherited together from a single parent. These are single nucleotide polymorphisms that tend to inherit together over time and can serve as disease- gene markers. The examination of single chromosome sets (haploid sets), as opposed to the usual chromosome pairings (diploid sets), is important because mutations in one copy of a chromosome pair can be masked by normal sequences present on the other copy.
Monogenic inheritance:
Monogenic inheritance refers to the kind of inheritance, whereby a trait is determined by the expression of a single gene or allele, not by several genes as in polygenic inheritance.
Oligogenic inheritance:
A phenotypic outcome (physical characteristic or disease predisposition) that is determined by more than one gene.
Polygenic inheritance:
Polygenic inheritance refers to the kind of inheritance, which is determined by many genes, while oligogenic refers to those determined by a few genes.
Loci:
A locus (plural loci) is the specific location of a gene or DNA sequence on a chromosome. A variant of the DNA sequence at a given locus is called an allele.
Alleles:
An allele is an alternative form of a gene (one member of a pair) that is located at a specific position on a specific chromosome. These DNA codings determine distinct traits that can be passed on from parents to offspring.
Codon:
Three bases in a DNA or RNA sequence which specify a single amino acid.
Nucleotide:
A nucleotide is an organic molecule made up of a nucleotide base, a five-carbon sugar (ribose or deoxyribose) and at least one phosphate group. Nucleotides make up the basic units of DNA and RNA molecules.
Genetic marker:
A gene or (a fragment of) DNA sequence having a known location on a chromosome, have an easily identifiable phenotype and whose inheritance pattern can be followed.
Candidate gene:
Candidate gene is a gene whose chromosomal location is associated with a particular disease or another phenotype. Because of its location, the gene is suspected of causing the disease or another phenotype. Association studies with candidate genes have been widely used for the study of complex diseases.
Genetic map:
It is a chromosome map of a species that shows the position of its known genes and/or markers relative to each other, rather than as specific physical points on each chromosome.
Susceptibility genes:
A gene mutation that increases an individual’s susceptibility or predisposition to a certain disease or disorder is called as a susceptibility gene.
Homozygous:
Possessing two identical forms of a particular gene, one inherited from each parent.
Heterozygous:
Possessing two different forms of a particular gene, one inherited from each parent.
Linkage:
The tendency of genes to be inherited together as a package because of their location near one another on the same chromosome.
Linkage disequilibrium:
Occurrence of two linked alleles at a frequency higher or lower than expected in a population, on the basis of the gene frequencies of the individual genes.
Mutation:
It is a relatively permanent change in hereditary material involving either a physical change in chromosome relations or a biochemical change in the codons that make up genes.
Polymorphism:
When a specific allele occurs in at least 1% of the population, it is said to be a genetic polymorphism.
Single nucleotide polymorphism (SNP):
This is the simplest type of polymorphism, which results from a single base mutation which substitutes one nucleotide for another.
Restriction fragment length polymorphism (RFLP):
Digestion of a piece of DNA containing the relevant site with an appropriate restriction enzyme can distinguish alleles or variants based on the resulting fragment sizes via electro-phoresis. This is called as “Restriction fragment length polymorphism” (RFLP).
Variable number of tandem repeats (VNTRs or “mini-satellites”):
A polymorphism that results from the deletion or insertion of a section of DNA in the existence of variable numbers of repeated base or nucleotide patterns in a genetic region. These repeated base patterns range in size up to several hundreds of base pairs, known as “variable number of tandem repeats” (VNTRs or “minisatellites”).
Simple tandem repeats (STRs) or Microsatellite repeat polymorphism:
This is the polymorphism resulting from two, three, or four nucleotides repeated over some variable number of times.
Research done on genetic aspect of periodontal diseases
A lot of research has been done to find out the association of genetic markers and the severity of periodontal diseases. For the ease of understanding, let us divide research into five categories according to the method used for studying the association of genetics and severity of periodontal diseases,
- The study of inherited diseases and genetic syndromes.
- Family studies.
- Twin studies.
- Population studies.
- Single nucleotide polymorphisms (SNP).
Study of inherited diseases and genetic syndromes
Papillon‐Lefèvre syndrome:
It is a condition in which the cardinal clinical features are severe periodontitis and palmar-plantar hyperkeratosis with variation in the severity and extent 5. Genetic linkage analysis on chromosome 11q14 and mutational analysis have permitted the identification of mutations in the cathepsin C gene in patients with Papillon‐Lefèvre syndrome 6, 7. Due to mutations of this gene, there is a loss of protease activity of the cathepsin C protein.
Haim-Munk syndrome:
Both Papillon‐Lefèvre syndrome and Haim-Munk syndrome are allelic variants of mutations of the cathepsin C gene. Along with palmer planter hyperkeratosis, Haim-Munk syndrome has additional features like arachnodactyly, acro-osteolysis, atrophic changes of nails and deformity of fingers.
Chediak-Higashi syndrome:
It is a rare disease transmitted as an autosomal recessive trait. In this condition, defect lies with polymorphonuclear leukocyte (PMN’s). A structural defect resulting from the fusion of azurophilic and specific granules into giant granules, called “megabodies,” is the characteristic of neutrophils from individuals with this disease. Polymorphonuclear leukocyte ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Acatalasia:
It is a rare hereditary metabolic disorder caused due to lack of enzyme catalase.
Hypophosphatasia:
This condition is caused due to a mutation in genes coding for tissue non-specific alkaline phosphatase (TNSALP). The TNSALP gene is located on chromosome 1p36.19 and consists of 12 exons distributed over 50 kb. This condition is characterized by defective bone and teeth mineralization and deficiency of serum and bone alkaline phosphatase activity. Depending upon the age of the patient at the time of diagnosis, this condition has been recognized in five forms: prenatal (lethal), infantile, childhood, adult, and odontophosphatasia.
Prenatal (lethal) form:
In this form, there is markedly impaired mineralization in utero because of which the fetus has skin-covered osteochondral spurs protruding from the forearms or legs (diagnostic marker). The infant survives for a few days but dies due to respiratory complications which are hypoplastic lungs and rachitic deformities of the chest.
Infantile form:
The patient is normal at birth and clinical features appear during the first 8 months. The patient has rachitic deformities, premature craniosynostosis (result-ing in increased intracranial pressure). Other findings include hypercalcemia, hypotonia, polydipsia, polyuria and dehydration. There is a premature loss of deciduous teeth.
Childhood form:
Clinical symptoms are a dolicho-cephalic skull and enlarged joints, a delay in walking, short stature and waddling gait. Dental findings include premature loss of primary dentition.
Adult form:
Patient presents with stress fractures of the metatarsals, thigh pain due to pseudofractures of the femur and premature loss of deciduous dentition.
Odontophosphatasia:
This condition is primarily characterized by dental abnormalities. These include, premature exfoliation of fully rooted primary teeth and/or severe dental caries. Radiographic investigations show enlarged pulp chambers and reduced alveolar bone.
Ehlers-Danlos syndrome:
It comprises of a collection of connective tissue disorders characterized by defective collagen synthesis. Individuals with defective collagen Types IV and VIII, inherit this defect in an autosomal dominant manner and have an increased susceptibility to periodontitis 10, 11.
Hyper-IgE (Job’s syndrome, HIE):
This clinical condition is characterized by a marked elevation of IgE. The defect has been possibly localized to chromosome 7q21. The condition is clinically characterized by chronic dermatitis (eczematoid rash). The patient has serious, lifelong bouts of recurrent infections by opportunistic organisms (Staphylococcus aureus and Candida albicans) which result in skin abscesses, remarkable in their lack of erythema (“cold” abscesses). Oral and periodontal findings are periodontitis with oral ulcerations.
Weary-Kindler syndrome:
In this syndrome abnormalities of the epidermal keratinocytes is seen 12. Clinical manifestations of this condition also include epidermolysis bullosa and poikiloderma congenitale. Periodontal manifestations include early-onset periodontitis 12, 13. The abnormality associated with this syndrome lies with the basement membrane, particularly in the expression of collagen Type VII 13.

Leukocyte adhesion deficiency type I and Leukocyte adhesion deficiency type II:
A detailed description of leukocyte adhesion deficiency type I and type II has been given in “Role of neutrophils in host-microbial intractions”.
Family studies
If the disease has a genetic basis, it is passed from parents to children in a predictable manner, and usually, segregate in families, as predicted by Mendel’s laws 14. Mendelian mode of inheritance includes autosomal dominant, autosomal recessive or X-linked inheritance. According to Mendelian laws, the disease phenotype is manifested over a wide range of environments, and although environmental factors and other genes can modify the clinical presentation, in most cases the mutation will manifest in a remarkably similar phenotype. The study of passage of genes in families from parents to siblings is known as ‘segregation analysis’.
Segregation analysis are applied to determine whether a trait transmission appears to fit the Mendelian or another mode of genetic transmission. When the genetic models of transmission are compared, important information about the mode of transmission (e.g. autosomal, X-linked, dominant, recessive, complex, ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
An X-linked inheritance of aggressive periodontitis (Grade C periodontitis) was proposed by Melnick et al. (1976) 16 based on a preponderance of female probands. The female: male ratio of affected persons was approximately 2:1. In another study, an autosomal recessive mode of inheritance was proposed in the Finnish population, whereas an autosomal dominant mode of inheritance was proposed in African American and Caucasian families 17. In a segregation analysis, Schenkein (1998) 18 theorized that aggressive periodontal disease and IgG2 responsiveness to bacterial lipopolysaccharide segregate independently as a dominant and codominant trait, respectively. He proposed that subjects with only one copy of IgG2 allele would develop severe periodontal disease as compared to subjects with 2 copies of high IgG2 response allele with one aggressive periodontal disease allele.
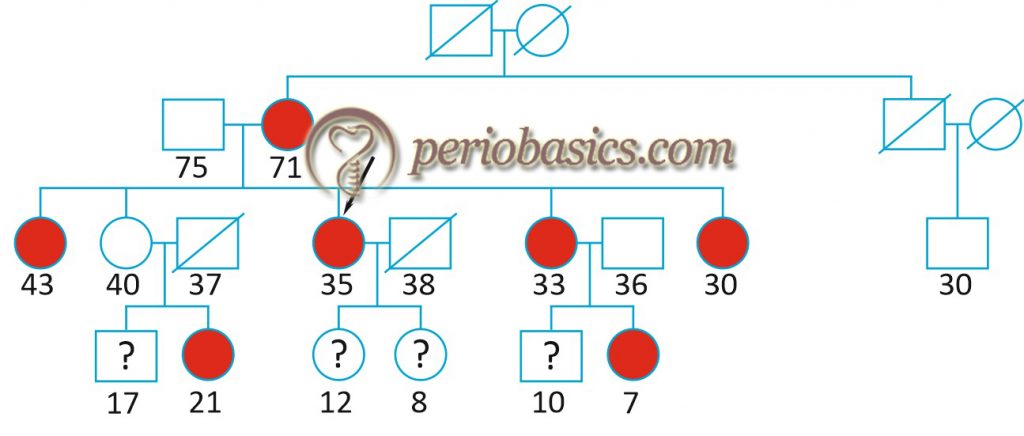
Hence, it can be concluded from the above findings that genetics plays a role in the inheritance of Grade C periodontitis and a few loci, each with relatively small effects contributes to the development of the disease with or without interaction with the environmental factors.
Linkage analysis
The linkage analysis studies are based on the fact that alleles at syntenic gene loci in close proximity on the same chromosome tend to be passed together from generation to generation (i.e. segregate), as a unit. The genes violate Mendel’s law of independent assortment. These genes are referred to as linked. Boughman et al. (1985) 19 were first to demonstrate the link-age between aggressive periodontitis (Grade C periodontitis) and dentinogenesis imperfecta. A specific chromosomal region (4q11-13) associated with aggressive periodontitis was identified by the authors, which was near the gene for dentinogenesis imperfect. Li Y et al. (2004) 20 reported ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
Twin studies
Twin studies provide important information regarding the genetic influences in disease and for partitioning the relative contribution of genes and environment to a trait. Monozygous twins arise from one fertilized egg, whereas dizygous twins arise from two different eggs and two different sperms. Monozygous twins are genetically identical, whereas dizygous twins are only as genetically similar as brothers and sisters would be, on an average, they share ~50% of their genes in common. Any difference between disease expression and progression in monozygous twins is due to environmental factors, whereas disease discordance between dizygous twins could arise from both environmental and genetic differences. This difference can be used to estimate the relative contribution of genes (heredity) and environmental factors in the expression and progression of a disease.
Some very informative twin studies are discussed here. Corey et al. (1993) 3 did a study on 4908 twin pairs and found that 9% of subjects, consisting of 116 identical and 233 nonidentical twin pairs, reported a history of periodontitis. Their study suggested that heritable factors are important in the reported periodontitis experience. The drawback of this study was that environmental factors like smoking were not taken into consideration because of which the results could be biased. Another study was done by Michalowicz et al. (1991) 4 on monozygotic and dizygotic twins. In this study, dizygous twins reared apart (dizygous-A) and reared together (dizygous-T) and monozygous twins reared apart (monozygous-A) and reared together (monozygous-T) were examined for the mean ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Population studies
Studies done on different populations provide important information about the genetic component of a disease. Studies done on populations have revealed that the polymorphisms in the interleukin (IL)-1 gene cluster linked with periodontitis 22 are found in approximately 30% of the European population. However, the prevalence is dramatically lower in Chinese (2.3%) and thus the usefulness of the ‘composite genotype’ of allele 2 of both interleukin (IL)-1α (+ 4845) and interleukin (IL)-1β (+ 3954) for determining susceptibility in Chinese patients is dubious 23. So, we can expect variations in the expression of disease in different populations.
Single nucleotide polymorphisms in periodontal disease
Genetic predisposition of certain individuals towards periodontal breakdown has been well documented. Single nucleotide polymorphism in cytokines, HLA, immuno-receptors, proteases and some structural molecules has been well investigated. These can be classified as follows,
Cytokine polymorphism,
- IL-1.
- TNF-α.
- IL-4.
- IL-6.
- IL-10.
Fc γ receptor (FcγR) gene polymorphisms,
- FcγRIIa 131 polymorphism.
- FcγRIIIa 158 polymorphism.
- Bi-allelic polymorphism in the FcγRIIIb.
Antigen-recognition related gene polymorphism,
- Human leukocytic antigen polymorphism.
Metabolism-related gene polymorphism,
- Polymorphisms in the vitamin D receptor (VDR) gene.
Polymorphisms in the CD14 Gene.
Polymorphisms in the pattern recognition receptor genes.
Proteases polymorphism,
- Matrix metalloproteinases (MMP) polymorphisms.
Miscellaneous gene polymorphisms,
- Cathepsin C gene polymorphism.
Let us now discuss these polymorphisms in detail,
Cytokine polymorphism:
Cytokines play an important role in a number of different physiological processes, but if expressed inappropriately, they lead to the connective tissue destruction. Pro-inflammatory cytokines include IL-1α, IL-1β, IL-6, IL-8 and TNF-α whereas anti-inflammatory cytokines include IL-4, IL-10, IL-13 and transforming growth factor (TGF)-β. Under pathologic conditions such as those that occur in periodontal disease, the balance between pro- and anti-inflammation is directed towards proinflammatory activity 24.
IL-1 polymorphism:
The three cytokines originally described as the members of the IL-1 family are IL-1α and IL-1β, which have agonist activity, and IL-1Ra, a physiological antagonist to other IL-1 cytokines. They are secreted by a variety of cell types including monocytes, macrophages, dendritic cells, epithelial cells, keratinocytes, and fibroblasts. IL-1α is a regulator of intracellular events and a mediator of local inflammation, whereas Il-1β is primarily an extracellular protein, released from the cells 25. Genes that regulate the production of IL-1 are located on chromosome 2q13. Genes IL1A and IL1B control the production of the pro-inflammatory proteins, IL-1α, and IL-1β, respectively. IL1RN controls the synthesis of the antagonist protein (IL-1Ra) 26.
A study by Kornman et al. (1997) 22 demonstrated that the occurrence of IL-1A (-889) and\IL-1B (+3954) polymorphisms simultaneously was associated with the severity of chronic periodontitis in non-smoker Caucasians. This composite genotype involves mutation -889 G→T (in linkage disequilibrium with 4845) in the IL-1α gene and mutation 3953 C→T (previously numbered 3954) in the in the IL-1β gene 27. It was reported that patients who were having this periodontitis associated genotype (PAG) were seven times more prone to have severe periodontitis as compared to patients who were negative for PAG 22. Other studies also demonstrated that either the smoking status or the IL-1 PAG contributes to the severity of periodontitis 28, 29.
In a study, it was demonstrated that low levels of bacterial load in individuals with mutated composite IL-1 genotype was able to induce the same amount of periodontitis as compared to mutation-negative individuals 30. While comparing the antibody response it was demonstrated that PAG-positive patients had a lower systemic immunoglobulin G response to periodontal microbiota than PAG-negative patients 31. One study done on Caucasian population investigated the distribution of polymorphisms in the IL-1 gene family among periodontitis patients and controls where both smoking and microbiological parameters including the presence of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans were also taken into consideration. The results showed a higher frequency of allele 2 carriage in IL-1A (-889) and IL-1B (+3954) single ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
On the contrary, some studies did not find any difference between the IL-1β production from monocytes from ‘composite genotype’ positive and negative patients. In one study 34 peripheral blood monocytes from ‘composite genotype’ positive and negative patients were examined to find out whether the IL-1β polymorphism was correlated with increased IL-1β expression by monocytes in response to a periodontal bacterial stimulus. The workers found no significant differences in IL-1β production in response to any stimulant tested.
So, conclusions drawn from the above discussion are 35:
- IL-1β polymorphism is a useful periodontal disease marker in defined populations only.
- The composite polymorphisms may be a part of several factors involved in the genetic risk for Grade C periodontitis. However, confirmation of the functional significance of this gene polymorphism remains to be confirmed.
TNF-α polymorphism:
TNF-α is a potent immunomodulator and proinflammatory cytokine that has been implicated in the pathogenesis of the autoimmune and infectious diseases. It induces the secretion of collagenase by fibroblasts, stimulates the resorption of cartilage and bone, and has been implicated in the destruction of periodontal tissue in periodontitis 36. Genes for TNF-α lies on the short arm of chromosome 6 (6p21.3) within the class III region of the major histocompatibility complex. As TNF-α acts synergistically with IL-1, higher production of IL-1 and TNF-α has been associated with enhanced response to infection. SNPs in the gene encoding TNF- α are mainly studied in the promoter region at positions -1031, -863, -367, -308, -238 but also in the coding region in the first intron at position +489. The single base transition polymorphism (G to A) at the -308 base site (TNF2) was found to result in a significantly higher transcription of the rare allele, TNF2, using a reporter gene construct 37. Other investigations have confirmed ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
IL-4 polymorphism:
It is a pleiotropic cytokine produced by the T helper 2 cell subpopulation. The gene encoding for IL-4 are located on chromosome 5q31.1. It is the major cytokine responsible for B-lymphocyte mediated immunity. It has got anti-inflammatory properties. The most studied polymorphisms in IL-4 are IL-4 -590 promoter polymorphism and 70-bp VNTR polymorphism. One study done on Caucasian patients with aggressive periodontitis showed that 27.8% of them had these polymorphisms 41. Most of the studies conclude that there is no evidence for any association between single SNPs of IL-4 and chronic periodontitis 42, 43, however in one study, haplotype T(-590)/T(-33)/allele 2 VNTR (70 base pairs)2 of the IL-4 gene polymorphism was shown to be significantly more frequent in patients with chronic periodontitis than in healthy individuals (17.0% in cases vs. 11.0% in controls; odds ratio 1.85) 44.
IL-6 polymorphism:
Genes coding for IL-6 are located on chromosome 7p21. It is synthesized primarily by monocytes, macrophages, and other antigen-presenting cells and has pro-inflammatory and hematopoietic activities. Many studies have demonstrated the association between periodontal diseases and IL-6 polymorphism at position -174 1, 45, 46.
IL-10 polymorphism:
Gene encoding for IL-10 are located on chromosome 1q31-q32, in a cluster with closely related interleukin genes, including IL-19, IL-20, and IL-24. It is produced by a variety of cells, which include monocytes/macrophages, dendritic cells, B-lymphocytes (particularly the CD5+ B cells), various subsets of CD4+ and CD8+ T-cells 47 and also by human ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
Fc γ receptor (FcγR) gene polymorphisms:
Leukocytes derived from both the myeloid and lymphoid lineages express receptors (FcγR) for the constant (Fc) region of immunoglobulin G molecules 53. Genes for Fcγ receptor are located on chromosome 1 and they encode for three main receptor classes: FcγRI (CD64), FcγRII (CD32) and FcγRIII(CD16). These classes are further subdivided into subclasses: FcγRIa and b, FcγRIIa, b and c, and FcγRIIIa and b. FcγRIIa is found on all granulocytes, antigen-presenting cells, platelets, endothelial cells and a subset of T-cells. FcγRIIIa is present on monocytes and macrophages, NK cells and a subset of T-cells. The FcγRIIIb is the major IgG receptor of neutrophils 54. The interaction between Fcγ receptors and IgG triggers a variety of biological responses, including phagocytosis, endocytosis, antibody-dependent cellular cytotoxicity, the release of inflammatory mediators, and enhancement of antigen presentation 55.
FcγRIIa 131 polymorphism:
In this polymorphism, there is a transition of G to A in the FcγRIIa gene, which results in the substitution of histidine (H) (N-allele) for arginine (R) (R-allele) at amino acid position 131 of the receptor. FcγRIIa-H131 binds IgG2 immune complexes efficiently, whereas, the FcγRIIa-R131 allotype cannot mediate this interaction 56. The carriage rate of the R-allele is relatively high in Caucasians and African-Americans 54, 57, 58. Studies show that ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
FcγRIIIa 158 polymorphism:
In this case, there is G to T transition in the FcγRIIIa gene, which results in an amino acid 158-valine (V) (N-allele) substitution for 158-phenylalanine (F) (R-allele). FcγRIIIa- V158 has a higher affinity for IgG1 and IgG3 than FcγRIIIa-F158 and FcγRIIIa-V158 can bind IgG4, while FcγRIIIa-F158 cannot. The carriage rate of the FcγRIIIa-F158 allele is lower in Japanese than in Caucasians and African- Americans. One study has demonstrated that FcγRIIIa N-allele (V158) is a putative risk factor for periodontitis, in particular for aggressive periodontitis in a group of Dutch patients 54.

Bi-allelic polymorphism in the FcγRIIIb:
In neutrophils, FcγRIIIb exists in two allelic forms, NA1, and NA2. This polymorphism is a result of nucleotide substitutions resulting in changes in four amino acids, which consequently results in differences in glycosylation. FcγRIIIb-NA2 type binds less efficiently human IgG1 and IgG3 immune complexes than FcγRIIIb-NA1. A study done on Japanese population showed that FcγRIIIb R-allele (NA2) was associated with generalized (G)-EOP 59. In African-Americans, an association between the FcgRIIIb and periodontitis has been found.
Antigen – recognition related gene polymorphism:
Human leukocyte antigen (HLA) polymorphism:
The HLA complex is one of the most important components of the host response. A lot of investigations have been done on polymorphism in HLA molecules. One study done on HLA-DR molecules in patients with periodontitis found a significant association between several DRB1 alleles and the disease 62, but another study 63 found no association between the presence of HLA-DQB1 in European Caucasians and the occurrence of early-onset periodontitis.
Metabolism-related gene polymorphism
Polymorphisms in the VDR gene:
As it is well known that vitamin D plays an important role in calcium and phosphorus metabolism, the polymorphism in vitamin D receptor gene can play a role in periodontal diseases. The human VDR gene is located on chromosome 12q12–q14, with a cluster of polymorphisms associated with BsmI, ApaI and TaqI sites. In one study, restriction fragment length polymorphism (RFLP) for TaqI in exon 9 in 69 Caucasian patients with aggressive periodontitis and 72 race-matched controls was studied. Results demonstrated a higher prevalence of TaqI RFLP (t) in the patients with localized aggressive periodontitis than in the controls 64. Whereas, another study ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
Polymorphisms in the CD14 Gene:
It is found in two distinct forms: membrane CD14 (mCD14), expressed primarily on the surface of monocytes/ macrophages and neutrophils, and a soluble form (sCD14) that lacks the glycosylphosphatidylinositol anchor 67, 68. Soluble CD14 mediates the response to lipopolysaccharide (LPS) in cells lacking membrane-bound CD 14, such as endothelial and epithelial cells 68. Polymorphisms in the CD14 gene have been studied in the promoter region at positions -159, -1619, -1359, -1145 and -809. One study done on Czech patients demonstrated that the -1359 C/T genotype polymorphism was associated with the severity of chronic periodontitis, while the -159 G/T genotype was not 69. Another study investigated the association between an SNP at position -159 in the promoter region of the CD14 gene with CD14 expression and TNF-α production by monocytes. Results demonstrated that the polymorphism C-159-T resulted in an 80% increase in constitutive gene expression 70.
Polymorphisms in the pattern recognition receptor genes:
The pathogen-associated molecular patterns (PAMPs) that are expressed on microorganisms are recognized by the innate immune system of the host. Extra- and intracellular receptors like CD14, CARD15, and Toll-like receptors (TLRs) recognize PAMPs of Gram-positive and Gram-negative bacteria and mediate the production of cytokines necessary for further development of effective immunity. Both TLR2 and TLR4 use CD14 as a co-receptor. Polymorphism in CD14 genes has already been discussed. As far as TLR2 and TLR4 are concerned, their genes have been located on chromosome 4q32 and 9q32-q33, respectively. Polymorphisms studied for TLR2 are Arg677Trp and Arg753Gln. These polymorphisms lead to hampering of the ability of TLR2 to mediate the response to bacterial components 71. Polymorphisms reported for TLR4 are Asp299Gly and Thr399Ile. These lead to ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Proteases polymorphism:
Matrix metalloproteinases (MMP) polymorphisms:
In MMP’s a single nucleotide polymorphism in the promoter region -1607 base pair of MMP-1 gene, 5′-GGA-3′, instead of 5′-GAT-3′ has been found to be associated with increased risk of generalized Grade C periodontitis 75. One study done on patients with Brazilian background showed that the MMP-1 (-1607) 2G/2G genotype was more frequently observed in 26 patients with severe chronic periodontitis than in 24 patients with moderate chronic periodontitis and 37 controls 76. Another study showed that three MMP-1 polymorphisms (-1607 1G/2 G, -519 A/G, and -422 A/T) showed only a small effect on Grade A/B periodontitis in the Czech population 77. A study done on Japanese population demonstrated that MMP-1 and⁄or MMP-3 single nucleotide polymorphisms were not associated with susceptibility to periodontitis 78.
Miscellaneous gene polymorphisms:
Cathepsin C gene polymorphism:
Cathepsin C functions as a central coordinator for the activation of many serine proteases in inflammatory cells. It has been recognized that cathepsin C is responsible for neutrophil recruitment and production of chemokines and cytokines in many inflammatory diseases. The cathepsin C gene (11q14.2) spans 4.7 kb and has seven exons 6, 7. Cathepsin C gene mutations have been reported to be responsible for Papillon-Lefèvre Syndrome (PLS) 7 as well as similar conditions such as Haims-Munk Syndrome and juvenile periodontitis 79, 80. A detailed description of these diseases has been discussed in “Role of neutrophils in host-microbial interactions”.
Diagnosis of periodontal disease activity by genetic analysis
Genetic analysis can be used to detect the activity of genes at the active and inactive disease sites. In a study Demmer et al. (2008) 81 analyzed the whole genome to show the differential gene expression of healthy and diseased periodontal sites. The authors found thousands of up-regulated and down-regulated genes at the diseased sites as compared to healthy sites. These included genes for apoptosis, antimicrobial response, antigen presentation, regulation of metabolic process, signal transduction, and angiogenesis. However, the clinical application of this technique for the diagnosis of disease activity is limited.
Epigenetics and its role in periodontitis
Epigenetics is the study of changes in organisms caused by modification of gene expression rather than alteration of the genetic code itself. Epigenome refers to “epi” meaning outside the “genome.” It epigenetics, we study mitotically and meiotically heritable changes in gene function that are not dependent on DNA sequence. Epigenetic modifications include chemical alterations of DNA and associated proteins, leading to the remodeling of the chromatin and activation or inactivation of a gene. There are three types of epigenetic modifications 85, 86,
- DNA methylation,
- Histone modification, and
- RNA-associated silencing (micro-RNA).
Out of these three mechanisms, enzymatic DNA methylation of the C-5 position of cytosine residues in the CpG islands of the promoter region of a gene is considered as the most important epigenetic mechanisms in mammals 87. Let us discuss these mechanisms in detail.
DNA methylation:
In eukaryotes, it refers to the process by which a methyl group is covalently added to the carbon (at position 5) of cytosine in the DNA strand. Only those cytidine residues that are adjacent to guanidine i.e. the CpG sites (cytidine bound through a phosphate molecule to guanidine) in the DNA strand are targets for the methylation inducing enzymes. The DNA methylation is catalyzed by DNA methyl transferases (DNMT). There are 4 DNA methyl transferases,
- DNA methyl transferase1 (DNMT 1),
- DNA methyl transferase2 (DNMT 2),
- DNA methyl transferase3a (DNMT 3a), and
- DNA methyl transferase3b (DNMT 3b).
Methylation is an important step in gene transcription. Actively transcribed genes are characterized by the presence of CpG sites in their promoter regions that are hypomethylated. As the CpG sites in the promoter region are hypomethylated, it is possible for appropriate transcription factors to bind to their recognition sequence in the promoter region.

Hypermethylation of promoter region of genes is associated with transcriptional silencing of gene, thereby loss of gene expression. On the other hand, hypomethylation of promoter region of genes is associated with transcriptional activation of genes, thereby leading to gene expression.
Histone modification:
In the eukaryotic cells, the DNA is organized in a highly conserved structural polymer termed chromatin. The basic building block of chromatin is the nucleosome which consists of 146 bp of DNA wrapped around an octamer constituted of dimers of core histone proteins H2A, H2B, H3, and H4 held together by an H1 linker 88. The core histones are evolutionarily highly conserved proteins. They consist of globular domains with a covalently modifiable N-terminal tail at level of lysine and/or arginine residues. There are at least 9 different types of posttranslational modifications (such as acetylation, phosphorylation, methylation, biotinylation, SUMOylation, ADP ribosylation and ubiquitination) influence ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
RNA-associated silencing (micro-RNA):
MicroRNAs (miRNAs) are short, non-coding RNA molecules that mediate RNA silencing and regulate gene expression. miRNAs were discovered in 1993 and have been extensively studied ever since. miRNAs are responsible for changes in the cell epigenome because of their ability to modulate gene expression post-transcriptionally. These can directly target epigenetic factors, such as DNA methyltransferases or histone deacetylases, thus regulating chromatin structure. miRNAs predominantly regulate gene expression via translational inhibition, either by interfering with the ribosome assembly or by inducing its early dissociation 89-93. These miRNAs participate in the epigenetic mechanism by primarily three mechanisms,
- The expression of miRNAs is regulated by multiple epigenetic mechanisms,
- miRNAs can repress the expression of epigenetic factors; and
- miRNAs and epigenetic factors can cooperate to modulate common targets.
Hence, from the above discussion, we can conclude that the following epigenetic modifications are observed,
- DNA methylation- gene silencing.
- DNA demethylation- gene unsilencing.
- Histone acetylation- open chromatin, active transcription.
- Histone methylation- closed chromatin, silencing of transcription (generally).
- DNA methylation and miRNAs cooperate in the suppression of gene expression and protein translation of common targets.
- miRNAs control the chromatin structure by affecting the “histone code” and targeting key enzymes, known as histone modifiers.
Role of epigenetic changes in periodontal disease:
As already stated, the epigenetic changes affect gene expression by remodeling of chromatin and selective activation or inactivation of genes. These changes result in the changes in cytokine profile and immune mechanisms. There is a strong evidence in support of the role of genetic factors in the etiopathogenesis of periodontal diseases. However, the role of epigenetics in periodontics is a new area of research. The rate of disease progression, as well as response to periodontal treatment, may vary from patient to patients and this variability is influenced by genetic 21, as well as epigenetic factors 94. It has been found that in periodontitis, epigenetic modifications during inflammation occur locally at the biofilm-gingival interface around the teeth. Barros and Offenbacher (2014) 95 stated that ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Toll-like receptors (TLRs) play an important role in pathogen recognition. Hypermethylation and a decreased transcription of Toll-Like Receptor 2 (TLR 2) in periodontitis tissues have been reported 96. De Oliveira et al. (2011) 97 studied DNA methylation pattern in the TLR2 and TLR4 genes in gingival samples from healthy subjects, smokers, and non‐smokers affected by chronic periodontitis. The TLR4 gene promoter was unmethylated in most samples in all groups, whereas the TLR2 promoter was both methylated and unmethylated in most samples. Hypermethylation and subsequent low transcription of TLR2 in periodontitis tissues were reported 96. In a study on aggressive periodontitis patients, Baptista et al. (2014) 98 found an overall demethylation pattern of the suppressor of cytokine signaling 1 (SOCS1) gene. In healthy individuals, this demethylated level was higher, and also total demethylated samples were found higher for this group compared to periodontitis. A significant difference was also reported regarding the methylation pattern for the long interspersed element-1 (LINE-1) gene.
Epigenetic modifications have also been studied in the production of inflammatory cytokines in periodontitis. Studies have investigated DNA methylation of inflammatory cytokines in various forms of periodontitis. In one study, it was found that the IL-8 promoter was hypomethylated in oral epithelial cells from individuals with generalized aggressive periodontitis compared with healthy controls 99. These results were in agreement with another study where analysis of the IL-8 promoter indicated a tissue-specific pattern in DNA methylation 100. However, IL-6 promoter was found to be partially methylated in both healthy individuals and patients with periodontitis in spite of the fact that the expression of IL-6 was higher in patients with periodontitis 101. In contrast, Gomez et al. (2010) 102 ……. Contents available in the book……. Contents available in the book……. Contents available in the book……. Contents available in the book………
Can periodontal pathogens influence the epigenetic expression of cells of the periodontium? This question was addressed by Yin and Chung in 2011 105. The authors reported that Fusobacterium nucleatum stimulated gingival epithelial cells and these cells showed hypermethylation of the Mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1), thereby resulting in lack of Nuclear factor-kappa B production. On the other hand, stimulation of gingival epithelial cells with Porphyromonas gingivalis resulted in hypomethylation of ZNF287, a DNA binding protein believed to be involved in transcriptional regulation. Furthermore, P. gingivalis significantly decreased the tri-methylation of histone H3 K4 protein expression, but F. nucleatum did not. These findings supported the “stealth-like” properties of P. gingivalis supporting the concept of “keystone pathogens”.
Epigenetic therapy in the management of periodontitis:
Pharmacological agents can be used to revert the effects of epigenetic variations. Histone deacetylase inhibitors and DNA methyltransferase inhibitors have been investigated and are under research. Histone deacetylase inhibitors help in suppressing bone resorption by osteoclasts 106. Also, the deacetylase inhibitors help in promoting the osteoblast maturation 107. However, these treatment strategies still need a lot of research before they can be introduced into clinical practice.
Future of genetics in periodontics
A lot of research has been done to identify the genetic basis of periodontal diseases. The understanding of the genetics and periodontal disease progression has provided us valuable information for the identification of disease markers 82-84. Identification of candidate genes and their use as periodontal disease biomarkers is the potential clinical application of the research done so far in this field. These biomarkers may be helpful in identifying patients with enhanced disease susceptibility and associated systemic conditions, identifying sites with active disease and predicting sites that may have active disease in the future 82, 84. Because this is a relatively new field of research in periodontics, many new findings which relate genetics to periodontal diseases are expected in future.
Conclusion
Periodontal diseases are multi-factorial diseases. Genetics is an important part of these factors. Present knowledge suggests that genetic polymorphism is associated with the pathogenesis of periodontal diseases. Genetic factors, along with environmental factors are strongly associated with the development and progression of periodontal diseases. Identification of specific genes and genetic variants, aids in diagnosis and treatment of aggressive periodontal disease. In future, a lot of research is required in this direction to investigate different aspects of the genetic basis of periodontal diseases.
References
References are available in the hard-copy of the website.
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.